Elongation through transcription factors
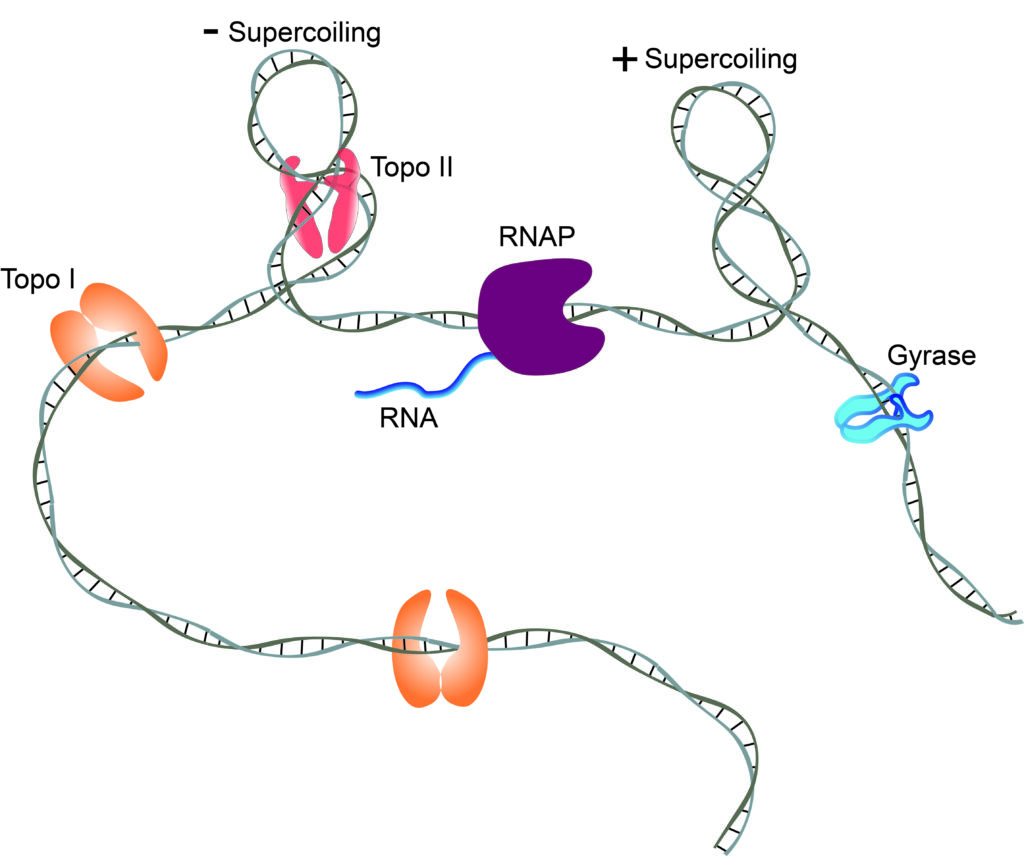
Understanding how RNA polymerases transcribe through transcription factors that decorate DNA without interfering with their physiological role is paramount to our understanding of maintenance of transcription regulation. We are particularly interested in roadblocks that alter topology by inducing DNA to wrap, kink or loop. Are these protein-induced DNA topological barriers strong roadblocks? Why, or why not? Using single molecule approaches, we observe E. coli RNAP traverse a protein roadblock, or entering and exiting a loop, and suggest mechanical pathways by which RNAP may do so.
Non canonical transcription termination
We are investigating alternative pathways to canonical termination where RNA polymerase releases the template, lets the DNA substrate go and diffuses in solution.
RNA polymerase I
RNA polymerase I (Pol I) is the eukaryotic polymerase that transcribes the genes that encode ribosomal RNA and is responsible for more than 60% of the cellular transcriptional activity. In collaboration with David Schneider (Univ. of Alabama, Birmingham), we characterized its elongation kinetics.
Topoisomerases
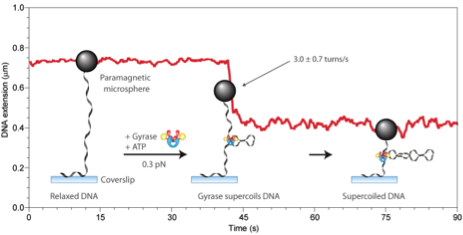
Topoisomerases are crucial in managing the level of DNA supercoiling largely arising from transcriptional activity of RNA polymerases. We study Type II topoisomerases using magnetic tweezers.
The experimental trace in the figure reveals the shortening of a DNA tether upon insertion of negative supercoils by Gyrase. The cartoons explain the different parts of the trace.
Relevant Publications
| Authors | Title | Journal | Volume | Pages | Year |
| Jin Qian, Bing Wang, Irina Artsimovitch, David Dunlap, Laura Finzi | “Force and the α-C-terminal domains bias RNA polymerase recycling” | Nature Communications | _ | _ | 2024 |
| Jin Qian, Allison Cartee, Wenxuan Xu, Yan Yan, Bing Wang, Irina Artsimovitch, David Dunlap, Laura Finzi | “Reciprocating RNA Polymerase batters through roadblocks” | Nature Communications | _ | _ | 2024 |
| Jin Qian, David Dunlap, Laura Finzi | “A Thermodynamic Model of Bacterial Transcription” | Physical Review E | _ | _ | 2022 |
|
Wenxuan Xu, Yan Yan, Irina Artsimovich, David Dunlap, Laura Finzi |
“Positive supercoiling favors transcription elongation through lac repressor-mediated DNA loops” | Nucleic Acids Research | _ | _ | 2022 |
|
Yue Lu, Gustavo Borjas, Cristine Hendrickson, Zsuzsanna Vörös, David Dunlap, Keith Shearwin, Laura Finzi |
“Proteins mediating different DNA topologies block RNAP elongation with different efficiency.” Editor’s choice-Journal cover. |
FEBS Letters | _ | _ | 2022 |
| Jin Qian, David Dunlap, Laura Finzi | “Basic mechanisms and kinetics of pause-interspersed transcript elongation” | Nucleic Acids Research | 49 | 15-24 | 2021 |
| Jin Qian, Wenxuan Xu, David Dunlap, Laura Finzi | “Single-molecule insights into torsion and roadblocks in bacterial transcript elongation” |
Transcription | 12 | 219-231 | 2021 |
|
Zsuzsanna Voros, Yan Yan, David D. Dunlap, Laura Finzi |
“Protein-mediated DNA looping enhances roadblocks” | Protein Science | _ | _ | 2017 |
|
S. Ucuncuoglu, K. L. Engel, P. K. Purohit, D. D. Dunlap, D. A. Schneider, Laura Finzi |
“Direct characterization of transcription elongation by RNA polymerase I” | PLOS One | _ | _ | 2016 |
Relevant Techniques
|
Method |
Used for |
|
AFM |
to visualize transcriptional elongation |
|
TPM |
to monitor the rate of transcriptional elongation and topological changes in the absence of force |
|
MTs |
to monitor the rate and dynamics of transcriptional elongation and topological changes in the presence of force |
|
C-Trap |
to monitor the dynamics of transcriptional elongation while simultaneously visualizing the activity of transcription factors |
Complete List of Published Work in MyBibliography:
https://www.ncbi.nlm.nih.gov/myncbi/browse/collection/40647244/?sort=date&direction=descending