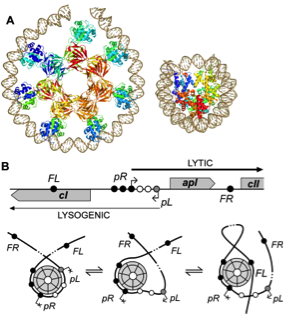

Figure adapted from H. Wang et al, NAR, 2013. Left: A – The 186 repressor is a heptamer of dimers which can wrap DNA. The nucleosome is shown besides it for scale comparison. B – (top) Schematic representation of the 186 DNA regulatory elements. pL is the lysogenic promoter from which the 186 repressor (CI) is transcribed; pR is the lytic promoter; solid black circles represent specific binding sites for 186 repressor DNA binding domains in the heptamer; empty circles represent non-specific binding sites. (bottom) Three possible 186 nucleoprotein conformations. Right: AFM analysis of the modes of interaction between 186 DNA and the 186 repressor protein. A – histogram showing the 186 Ci protein specific binding at the FL, pR and FR sites. B – AFM images showing the different nucleoprotein complexes detected and their frequency of occurrence. These different conformations play a role in the robustness of the 186 switch. C – histone octamer. D – lambda CI protein mediating a DNA loop.
Relevant Publications
| Authors | Title | Journal | Volume | Pages | Year |
| Yue Ding, Carlo Manzo, Geraldine Fulcrand, David Dunlap, Fenfei Leng and Laura Finzi | “DNA Supercoiling: a Regulatory Signal for the Lambda Repressor” | PNAS | 111(43) | 15402-15407 | 2014 |
| Marta Adelina Mendesa, Rosalinda Fiorella Guerra, Markus Christian Berns, Laura Finzi, Martin M. Kater and Lucia Colombo | “MADS-domain Transcription Factor Complex Induced Short-Range DNA Loop Formation is Essential for Target Gene Expression in Arabidopsis” | Plant Cell | 25 | 2560-2572 | 2013 |
| Haowei Wang, Ian Dodd, Keith Shearwin, David Dunlap and Laura Finzi | “Single molecule analysis of DNA wrapping and looping by a circular 14-mer of the 186 bacteriophage CI repressor” | Nucleic Acids Research | 41 | 5746-5756 | 2013 |
| Carlo Manzo, Chiara Zurla, David Dunlap and Laura Finzi | “The effect of non-specific binding of lambda repressor on DNA looping dynamics” | Biophys. J. | 103 | 1753-1761 | 2012 |
| Sachin Goyal, Chandler Fountain, David D. Dunlap, Fereydoon Family, Laura Finzi | “Stretching DNA to quantify non-specific binding” | PRE | 86 | 011905 | 2012 |
| Dale E.A. Lewis, Phuoc Le, Chiara Zurla, Laura Finzi, and Sankar Adhya | “Multi-level Auto-regulation of CI Protein in a λ lysogen” | PNAS | 108 (36) | 14807-14812 | 2011 |
| Paul Liebesny, Sachin Goyal, David Dunlap, Fereydoon Family and Laura Finzi | “Determination of the Number of Proteins Bound non-Specifically to DNA” | Journal of Physics: Condensed Matter | 22 | 414104 | 2010 |
| L. Finzi and D. Dunlap | “Single-molecule approaches to structure, kinetics and thermodynamics of transcriptional regulatory nucleoprotein complexes” | minireview – JBC | 285 | 18973-18978 | 2010 |
| C. Zurla, C. Manzo, D.D. Dunlap, D.E.A. Lewis, S. Adhya, L. Finzi | “Direct demonstration and quantification of long-range DNA looping by the λ bacteriophage repressor.” | NAR | 37 | 2789-2795 | 2009 |
| H. Wang, L. Finzi, D. Lewis and D. D. Dunlap | “AFM studies of the CI oligomers that secure DNA loops” | Curr. Pharmaceutical Biotechnology | 10 | 494-501 | 2009 |
| Suparna Sarkar-Banerjee, Sachin Goyal, Ning Gao, John Mack, Benito Thompson, David Dunlap, Krishnananda Chattopadhyay and Laura Finzi | “Specifically bound Lambda repressor dimers promote adjacent non-specific binding” | submitted |
Relevant Techniques
| Method | Used for |
| AFM | to image DNA-protein complexes and establish probabilities |
| TPM | to measure dynamics of loop formation |
| MTs | to measure how supercoiling affects genetic switches |
| FCS | to measure cooperative effects |
Complete List of Published Work in MyBibliography:
https://www.ncbi.nlm.nih.gov/myncbi/browse/collection/40647244/?sort=date&direction=descending

