DNA mechanical properties
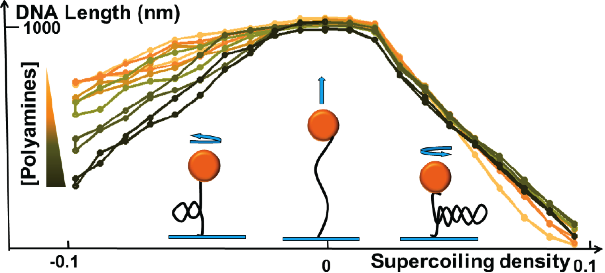
DNA bending and torsional elasticity directly affect most, if not all, genome functions by altering the affinity of proteins for their DNA binding sites, the likelihood of protein-protein or protein-DNA interactions, the activity of DNA enzymes and DNA/chromatin packaging. We study parameters that affect DNA elasticity such as polycations, base pairing, modified DNAs, DNA bending and coating proteins, histone post-translational modifications. The figure below shows how DNA torsional curves (hat curves) obtained with magnetic tweezers can be used to study the effect of polycations on DNA torsional elasticity.
DNA topology and phase changes
DNA may undergo topological changes depending on the environment. For example, it may be condensed or extended, right- or left-handed. These different forms and the partition of DNA between them is relevant to diseases and forces in the cell play a significant role in shifting the equilibrium between these forms and phases.
Liquid-liquid condensates
This is a new area of interest in the Finzi-Dunlap group and new projects are in progress.
Publications
| Authors | Title | Journal | Volume | Pages | Year |
| Wenxuan Xu, Yan Yan, Irina Artsimovich, David Dunlap and Laura Finzi | “Positive supercoiling favors transcription elongation through lac repressor-mediated DNA loops” | Nucleic Acids Research | 50 | 2826–2835 | 2022 |
| Wenxuan Xu, Laura Finzi and David Dunlap | “Energetics of twisted DNA topologies” | Biophysical Journal | 120 | 3242–3252 | 2021 |
| Alexander Zhang, Yan Yan, Fenfei Leng, David Dunlap, and Laura Finzi | “Ionic strength modulates HU protein-induced supercoiling” | BIORXIV/2021/464438 | – | – | 2021 |
| Yan Yan, Wenxuan Xu, Sandip Kumar, Alexander Zhang, Fenfei Leng, David D. Dunlap and Laura Finzi | “Negative DNA supercoiling makes protein-mediated looping deterministic and ergodic within the bacterial doubling time” | Nucleic Acids Research | 49(20) | 11550–11559 | 2021 |
| Domenico Salerno, Francesco Mantegazza, Valeria Cassina, Matteo Cristofalo, Qing Shao, Laura Finzi, David Dunlap | “Nanomechanics of negatively supercoiled diaminopurine-substituted DNA” | Nucleic Acids Research | 49(20) | 11778–11786 | 2021 |
| M. Cristofalo, D. Kovari, R. Corti, D. Salerno, V. Cassina, D. Dunlap, F. Mantegazza | “Nanomechanics of diaminopurine-substituted DNA: Third hydrogen-bond stabilizes base pair interactions but facilitates conformational changes” | Biophysical Journal | – | – | 2019 |
| Yan Yan, Yue Ding, David Dunlap and Laura Finzi | “Protein-mediated loops in supercoiled DNA create large topological domains” | Nucleic Acids Research | 46(9) | 4417–4424 | 2018 |
| Yan Yan, Fenfei Leng, Laura Finzi and David Dunlap | “Protein-mediated looping of DNA under tension requires supercoiling” | Nucleic Acids Research | 46(5) | 2370–2379 | 2018 |
| Laura Finzi and David Dunlap | “Supercoiling biases the formation of loops involved in gene regulation” | Biophysical Reviews | 8(1) | 65–74 | 2016 |
| Laura Finzi and Wilma Olson | “The Emerging role of DNA supercoiling as a dynamic player in genomic structure and function” | Biophysical Reviews | 8(1) | 1–3 | 2016 |
| Monica Fernandez, Qing Shao, Chandler Fountain, Laura Finzi, David D. Dunlap | “E. coli gyrase fails to supercoil diaminopurine-substituted DNA negatively” | JMB | 427 | 2305–2318 | 2015 |
| Yue Ding, Carlo Manzo, Geraldine Fulcrand, David Dunlap, Fenfei Leng and Laura Finzi | “DNA Supercoiling: a Regulatory Signal for the Lambda Repressor” | PNAS | 111(43) | 15402–15407 | 2014 |
| Sachin Goyal, Chandler Fountain, David D. Dunlap, Fereydoon Family, Laura Finzi | “Stretching DNA to quantify non-specific binding” | PRE | 86 | 011905 | 2012 |
| Qing Shao, Sachin Goyal, Laura Finzi and David Dunlap | “Physiological levels of salt and polyamines favor writhe and limit twist in DNA” | Macromolecules | 45 | 3188−3196 | 2012 |
Relevant Techniques
| Method | Used for |
| TPM | to measure bending elasticity |
| MTs | to measure torsional elasticity and how bending and torsional elasticity depend on the tension applied to DNA or the chromatin fiber |
| to measure persistence length |
Complete List of Published Work in MyBibliography:
https://www.ncbi.nlm.nih.gov/myncbi/browse/collection/40647244/?sort=date&direction=descending

